Multilevel Modeling Solves the Multiple Comparison Problem: An Example with R
Multiple comparisons of group-level means is a tricky problem in statistical inference. A standard practice is to adjust the threshold for statistical significance according to the number of pairwise tests performed. For example, according to the widely-known Bonferonni method, if we have 3 different groups for which we want to compare the means of a given variable, we would divide the standard significance level (.05 by convention) by the number of tests performed (3 in this case), and only conclude that a given comparison is statistically significant if its p-value falls below .017 (e.g. .05/3).
In this post, we’ll employ an approach often recommended by Andrew Gelman, and use a multi-level model to examine pairwise comparisons of group means. In this method, we use the results of a model built with all the available data to draw simulations for each group from the posterior distribution estimated from our model. We use these simulations to compare group means, without resorting to the expensive corrections typical of standard multiple comparison analyses (e.g. the Bonferroni method referenced above).
The Data
We will return to a data source that we have examined previously on this blog: walking data from the Accupedo app on my phone. Accupedo is a free step-counting app that I’ve been using for more than 3 years to track how many steps I walk each day. It’s relatively straightforward to import these data into R and to extract the cumulative number of steps taken per hour.
In the data used for this post, I exclude all measurements taken before 6 AM (because the step counts are cumulative we don’t lose any steps by excluding these observations). This results in 18,117 observations of cumulative hourly step counts for 1113 days. The first observation is on 3 March 2015 and the last on 22 March 2018, resulting in just over 3 years of data!
A sample of the dataset, called aggregate_day_hour, is shown below:
| oneday | dow | hour | steps | week_weekend | |
|---|---|---|---|---|---|
| 1 | 2015-03-07 | Sat | 16 | 14942 | Weekend |
| 2 | 2015-03-07 | Sat | 17 | 14942 | Weekend |
| 3 | 2015-03-07 | Sat | 18 | 14991 | Weekend |
| 4 | 2015-03-07 | Sat | 19 | 15011 | Weekend |
| 5 | 2015-03-07 | Sat | 20 | 15011 | Weekend |
| 6 | 2015-03-07 | Sat | 21 | 15011 | Weekend |
| 7 | 2015-03-07 | Sat | 22 | 15011 | Weekend |
| 8 | 2015-03-07 | Sat | 23 | 15011 | Weekend |
| 9 | 2015-03-08 | Sun | 11 | 2181 | Weekend |
| 10 | 2015-03-08 | Sun | 12 | 2428 | Weekend |
Oneday represents the calendar year/month/day the observation was recorded, dow contains the name of the day of the week, hour is the hour of the day (in 24-hour or “military” style), steps gives the cumulative step count and week_weekend indicates whether the day in question is a weekday or a weekend.
Analytical Goal
Our ultimate purpose here is to compare step counts across a large number of days, without having to adjust our threshold for statistical significance. In this example, we will compare the step counts for 50 different days. The standard Bonferonni correction would require us to divide the significance level by the number of possible pairwise comparisons, 1225 in this case. The resulting significance level is .05/1225, or .00004, which imposes a very strict threshold, making it more difficult to conclude that a given comparison is statistically significant.
As mentioned above, the method adopted in this post is one that Andrew Gelman often advocates on his blog. This advice often comes in the form of: “fit a multilevel model!,” with a link to this paper by Gelman, Hill and Yajima (2012). I have to admit that I never fully understood the details of the method from the blog posts. However, by looking at the paper and the explanations and code presented in Chapter 12 of Gelman and Hill’s (2007) book, I think I’ve figured out what he means and how to do it. This post represents my best understanding of this method.1
In brief (see the paper for more details), we calculate a multilevel model using flat priors (e.g. the standard out-of-the-box model in the lme4 package). We extract the coefficients from the model and the standard deviation of the residual error of estimated step counts per day (also known as sigma hat), and use these values to simulate step count totals for each day (based on the estimated total step count and sigma hat). We then compare the simulations for each day against each other in order to make pairwise comparisons across days.
Sound confusing? You’re not alone! I’m still grappling with how this works, but I’ll gently walk through the steps below, explaining the basic ideas and the code along the way.
Step 1: Calculate the Model
In order to perform our pairwise comparisons of 50 days of step counts, we will construct a multi-level model using all of the available data. We’ll employ a model similar to one we used in a previous post on this blog. The model predicts step count from A) the hour of the day and B) whether the day is a weekday or a weekend. We specify fixed effects (meaning regression coefficients that are the same across all days) for hour of the day and time period (weekday vs. weekend). We specify a random effect for the variable that indicates the day each measurement was taken (e.g. a random intercept per day, allowing the average number of estimated steps to be different on each day). We center the hour variable at 6 PM (more on this below).
We can calculate this model using the lme4 package. We will also load the arm package (written to accompany Gelman and Hill’s (2007) book) to extract model parameters we’ll need to compute our simulations below.
# load the lme4 package
library(lme4)
# load the arm package
# to extract the estimate of the
# standard deviation of our residuals
# (sigma hat)
library(arm)
# center the hour variable for 6 pm
aggregate_day_hour$hour_centered <- aggregate_day_hour$hour -18
# compute the model
lme_model <- lmer(steps ~ 1 + hour_centered + week_weekend + (1 | oneday),
data=aggregate_day_hour)
# view the model results
summary(lme_model) The estimates of the fixed effects are as follows:
| Estimate | Std. Error | t value | |
|---|---|---|---|
| (Intercept) | 13709.05 | 143.67 | 95.42 |
| hour_centered | 1154.39 | 4.55 | 253.58 |
| week_weekendWeekend | -1860.15 | 268.35 | -6.93 |
Because we have centered our hour variable at 6 PM, the intercept value gives the estimated step count at 6 PM during a weekday (e.g. when the week_weekend variable is 0, as is the case for weekdays in our data). The model thus estimates that at 6 PM on a weekday I have walked 13,709.05 steps. The hour_centered coefficient gives the estimate of the number of steps per hour: 1,154.39. Finally, the week_weekendWeekend variable gives the estimated difference in the total number of steps per day I walk on a weekend, compared to a weekday. In other words, I walk 1,860.15 fewer steps on a weekend day compared to a weekday.
Step 2: Extract Model Output
Coefficients
In order to compare the step counts across days, we will make use of the coefficients from our multilevel model. We can examine the coefficients (random intercepts and fixed effects for hour and day of week) with the following code:
# examine the coefficients
coef(lme_model) Which the gives the coefficient estimates for each day (first 10 rows shown):
| (Intercept) | hour_centered | week_weekendWeekend | |
|---|---|---|---|
| 2015-03-03 | -184.31 | 1154.39 | -1860.15 |
| 2015-03-04 | 11088.64 | 1154.39 | -1860.15 |
| 2015-03-05 | 9564.42 | 1154.39 | -1860.15 |
| 2015-03-06 | 9301.65 | 1154.39 | -1860.15 |
| 2015-03-07 | 15159.48 | 1154.39 | -1860.15 |
| 2015-03-08 | 10097.27 | 1154.39 | -1860.15 |
| 2015-03-09 | 15163.94 | 1154.39 | -1860.15 |
| 2015-03-10 | 18008.48 | 1154.39 | -1860.15 |
| 2015-03-11 | 15260.7 | 1154.39 | -1860.15 |
| 2015-03-12 | 10892.19 | 1154.39 | -1860.15 |
We will extract these coefficients from the model output, and merge in the day of the week (week vs. weekend) for every date in our dataset. We will then create a binary variable for week_weekend, which takes on a value of 0 for weekdays and 1 for weekends (akin to that automatically created when running the multilevel model with the lme4 package):
# extract coefficients from the lme_model
coefficients_lme <- as.data.frame(coef(lme_model)[1])
names(coefficients_lme) <- c('intercept', "hour_centered", "week_weekendWeekend")
coefficients_lme$date <- row.names(coefficients_lme)
# create a day-level dataset with the indicator
# of day (week vs. weekend)
week_weekend_df <- unique( aggregate_day_hour[ , c('oneday', 'week_weekend') ] )
# join the week/weekend dataframe to the coefficients dataframe
library(plyr); library(dplyr)
coefficients_lme <- left_join(coefficients_lme,
week_weekend_df[,c('oneday', 'week_weekend')],
by = c("date" = 'oneday'))
# make a dummy variable for weekend which
# takes on the value of 0 for weekday
# and 1 for the weekend
coefficients_lme$weekend <- ifelse(coefficients_lme$week_weekend=='Weekend',1,0) Our resulting dataframe looks like this:
| intercept | hour_centered | week_weekendWeekend | date | week_weekend | weekend | |
|---|---|---|---|---|---|---|
| 1 | -184.31 | 1154.39 | -1860.15 | 2015-03-03 | Weekday | 0 |
| 2 | 11088.64 | 1154.39 | -1860.15 | 2015-03-04 | Weekday | 0 |
| 3 | 9564.42 | 1154.39 | -1860.15 | 2015-03-05 | Weekday | 0 |
| 4 | 9301.65 | 1154.39 | -1860.15 | 2015-03-06 | Weekday | 0 |
| 5 | 15159.48 | 1154.39 | -1860.15 | 2015-03-07 | Weekend | 1 |
| 6 | 10097.27 | 1154.39 | -1860.15 | 2015-03-08 | Weekend | 1 |
| 7 | 15163.94 | 1154.39 | -1860.15 | 2015-03-09 | Weekday | 0 |
| 8 | 18008.48 | 1154.39 | -1860.15 | 2015-03-10 | Weekday | 0 |
| 9 | 15260.7 | 1154.39 | -1860.15 | 2015-03-11 | Weekday | 0 |
| 10 | 10892.19 | 1154.39 | -1860.15 | 2015-03-12 | Weekday | 0 |
These coefficients will allow us to create an estimate of the daily step counts for each day.
Standard Deviation of Residual Errors: Sigma Hat
In order to simulate draws from the posterior distribution implied for each day, we need the estimated step counts per day (also known as y hat, which we’ll calculate using the coefficient dataframe above), and the sigma hat value (e.g. the standard deviation of the residual error from our estimated step counts) from the model. As Gelman and Hill explain in their book, we can simply extract the standard deviation using the sigma.hat command (this function is part of their arm package, which we loaded above). We’ll store the sigma hat value in a variable called sigma_y_hat. In this analysis, the standard deviation of the residual error of our estimated step counts is 2193.09.
# extract the standard deviation of the residual error of predicted step counts
# store in a variable called "sigma_y_hat"
sigma_y_hat <- sigma.hat(lme_model)$sigma$data
# 2913.093 Step 3: Sample 50 days and plot
For this exercise, we’ll randomly select 50 days for our pairwise comparisons. We’ll then plot the model estimated step count for the selected days. Here, I calculate the estimated step counts at 6 PM (using the formula and coefficients from the model) directly in the ggplot2 code.
# sample 50 random days
set.seed(1)
sampled_dates <- coefficients_lme[sample(nrow(coefficients_lme), 50), ]
# visualize estimated step count
# for each date
library(ggplot2)
ggplot(sampled_dates, aes(x = date, y = intercept + (week_weekendWeekend*weekend),
color = week_weekend)) +
# point plot
geom_point() +
# set the title
labs(title = "Model Estimated Step Count at 6 PM") +
# set the y axis title
ylab('Estimated Number of Steps at 6 PM') +
# turn off x axis title, make dates on x axis
# horizontal
theme(axis.title.x = element_blank(),
axis.text.x = element_text(angle = 90, vjust = .5)) +
# set the legend title
scale_colour_discrete(name ="Week/Weekend") Our sampled dates dataframe looks like this (first 10 rows shown):
| intercept | hour_centered | week_weekendWeekend | date | week_weekend | weekend | |
|---|---|---|---|---|---|---|
| 296 | 3985.94 | 1154.39 | -1860.15 | 2015-12-25 | Weekday | 0 |
| 414 | 16165.61 | 1154.39 | -1860.15 | 2016-04-21 | Weekday | 0 |
| 637 | 11963.4 | 1154.39 | -1860.15 | 2016-11-30 | Weekday | 0 |
| 1009 | 10950.75 | 1154.39 | -1860.15 | 2017-12-07 | Weekday | 0 |
| 224 | 14669.36 | 1154.39 | -1860.15 | 2015-10-14 | Weekday | 0 |
| 996 | 13273.51 | 1154.39 | -1860.15 | 2017-11-24 | Weekday | 0 |
| 1046 | 17883.73 | 1154.39 | -1860.15 | 2018-01-13 | Weekend | 1 |
| 731 | 10716 | 1154.39 | -1860.15 | 2017-03-04 | Weekend | 1 |
| 696 | 16334.55 | 1154.39 | -1860.15 | 2017-01-28 | Weekend | 1 |
| 69 | 15280.03 | 1154.39 | -1860.15 | 2015-05-12 | Weekday | 0 |
And our plot looks like this:
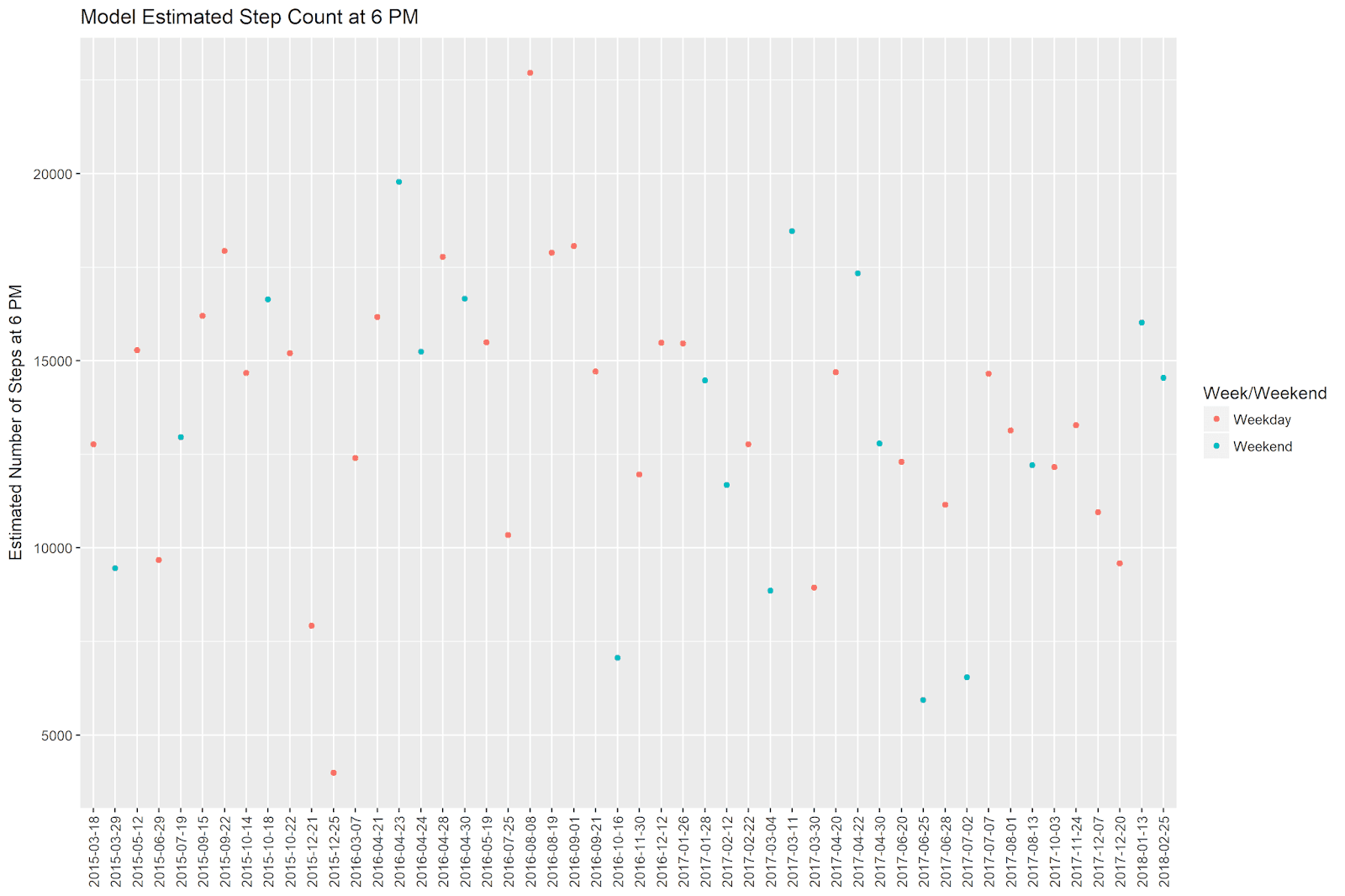
We can see that ‘2015-12-25’ (Christmas 2015) was day in which I walked very few steps. The highest step count is on ‘2016-08-08’, while the second-highest date is ‘2016-04-23’.
Note that the samples dataframe shown above is no longer in chronological order. However, ggplot2 is smart enough to understand that the x-axis is a date, and orders the values in the plot accordingly.
Step 4: Simulate Posterior Distributions of Step Counts Per Day
We now need to simulate posterior distributions of step counts per day. The method below is, as far as I can understand it, that applied in the Gelman et al. (2012) paper. I have adapted the logic from this paper and the Gelman and Hill book (particularly Chapter 12).
What are we doing?
Our model estimates the overall step counts as a function of our predictor variables. However, the model is not perfect, and the predicted step counts do not perfectly match the observed step counts. This difference between the observed and the predicted step counts for each day is called “residual error” - it’s what your model misses.
We will make 1000 simulations for each day with step count values that we did not actually see, but according to the model estimate and the uncertainty of that estimate (e.g. sigma hat), we could have seen. To do this, we simply calculate the model-predicted step count for each day, and use this value along with the sigma hat value (the standard deviation of our residual error) to sample 1000 values from each day’s implied posterior distribution.
How do we do it?
I wrote a simple function to do this. It takes our sample data frame and, for each row (date) in the dataset, it calculates the estimated step count using the regression formula and parameter estimates and passes this value, along with the sigma hat value, to the rnrom function, drawing 1000 samples from the implied posterior distribution for that day.
Note that I’m not including the hour_centered variable in this function. This is because I’m choosing to estimate the step count when hour_centered is zero (e.g. 6 PM because we centered on this value above).
# this function calculates the estimated step count
# and then draws 1000 samples from the model-implied
# posterior distribution
lme_create_samples <- function(intercept, w_wk_coef, w_wk_dum){
intercept_f <- intercept + (w_wk_coef * w_wk_dum)
y_tilde <- rnorm(1000, intercept_f, sigma_y_hat)
}
# here we apply the function to our sampled dates
# and store the results in a matrix called
# "posterior_samples"
posterior_samples <- mapply(lme_create_samples,
sampled_dates$intercept,
sampled_dates$week_weekendWeekend,
sampled_dates$weekend)
dim(posterior_samples)
# [1] 1000 50 Our resulting posterior_samples matrix has 1 column for each day, with 1000 samples for each day contained in the rows. Below I show a small subset of the entire matrix - just the first 10 rows for the first 5 columns. It is interesting to note that the first column, representing the first day (‘2015-12-25’) in our dataframe of samples above, has smaller values than the second column (representing the second day, ‘2016-04-21’). If we look at the intercepts in the samples dataset, we can see that the first day is much lower than the second. This difference in our model-estimated intercepts is propagated to the simulations of daily step counts.
| 3822.43 | 12517.38 | 10611.98 | 7940.35 | 15046.33 |
| 3532.09 | 13224.83 | 10720.97 | 5916.09 | 15879.66 |
| -298.5 | 18354.48 | 7002.13 | 7571.94 | 18489.92 |
| 2593.04 | 12354.25 | 8929.43 | 6891.52 | 9846.13 |
| 5203.44 | 17702.38 | 14202.8 | 8032.03 | 13051.09 |
| 7943.9 | 14611.36 | 9792.22 | 14878.74 | 9892.48 |
| 3686.51 | 15005.1 | 10546.27 | 5777.57 | 15511.05 |
| 5115.26 | 13865.52 | 10934.97 | 12029.68 | 21345.46 |
| 3829.2 | 15495.18 | 11781.92 | 11072.98 | 17209.84 |
| -25.57 | 18720.93 | 16681.49 | 10564.03 | 13591.19 |
Step 5: Compare the Simulations For Each Day
We can see in the above matrix of simulations that the first day (‘2015-12-25’, contained in the first column) has lower values than the second day (‘2016-04-21’, contained in the second column). In order to formally compare the the two columns, we will compare the simulations and see how many times the values in the first column are larger than those in the second column. If values in one column are larger more than 95% of the time, we will say that there is a meaningful (“significant”) difference in step counts between the two dates.
We will apply this logic to all of the possible pairwise comparisons in our sampled 50 dates. The following code accomplishes this (adopted from this tremendous Stackoverflow question + answer):
# do the pairwise comparisons
# first construct an empty matrix
# to contain results of comparisons
comparison_matrix<-matrix(nrow=ncol(posterior_samples),ncol=ncol(posterior_samples))
# give column and row names for the matrix
# (these are our observed dates)
colnames(comparison_matrix) <- sampled_dates$date
rownames(comparison_matrix) <- sampled_dates$date
# loop over the columns of the matrix
# and count the number of times the values
# in each column are greater than the values
# in the other columns
for(col in 1:ncol(posterior_samples)){
comparisons<-posterior_samples[,col]>posterior_samples
comparisons_counts<-colSums(comparisons)
comparisons_counts[col]<- 0 # Set the same column comparison to zero.
comparison_matrix[,col]<-comparisons_counts
}
# shape of output comparison matrix
dim(comparison_matrix)
# [1] 50 50 The first 10 rows of the first 5 columns of our comparison matrix look like this:
| 2015-12-25 | 2016-04-21 | 2016-11-30 | 2017-12-07 | 2015-10-14 | |
|---|---|---|---|---|---|
| 2015-12-25 | 0 | 998 | 970 | 954 | 995 |
| 2016-04-21 | 2 | 0 | 173 | 103 | 360 |
| 2016-11-30 | 30 | 827 | 0 | 395 | 722 |
| 2017-12-07 | 46 | 897 | 605 | 0 | 822 |
| 2015-10-14 | 5 | 640 | 278 | 178 | 0 |
| 2017-11-24 | 13 | 755 | 384 | 299 | 656 |
| 2018-01-13 | 3 | 514 | 173 | 114 | 379 |
| 2017-03-04 | 114 | 962 | 786 | 698 | 915 |
| 2017-01-28 | 7 | 660 | 273 | 223 | 529 |
| 2015-05-12 | 3 | 569 | 223 | 152 | 436 |
The counts in each cell indicate the number of times (out of 1000) that the column value is greater than the row value (we set the diagonals - comparisons of a day with itself - to zero in the above code). The comparison we identified above (‘2015-12-25’ and ‘2016-04-21’) is shown in the first row, second column (and the second row, first column - this matrix contains the same information but in reverse above and below the diagonals). The samples from ‘2016-04-21’ were greater than those from ‘2015-12-25’ 998 times out of 1000!
Step 6: Make the Pairwise Comparison Plot
We can make a heat map using ggplot2 to show which pairwise comparisons are “significantly” different from one another, akin to that used in Gelman et al. (2007).
Because ggplot2 requires data to be in a tidy format, we’ll have to melt the comparison matrix so that it has 1 row per pairwise comparison. The code to do this was based on these very clear explanations of how to make a heatmap with ggplot2:
# load the reshape2 package
# for melting our data
library(reshape2)
# melt the data
melted_cormat <- melt(comparison_matrix)
# rename the variables
names(melted_cormat)=c("x","y","count")
# identify which comparisons are "significant"
melted_cormat$meaningful_diff = factor(melted_cormat$count>950)
# and set the levels
levels(melted_cormat$meaningful_diff) = c('No Difference',
'Row > Column')
# sort the matrix by the dates
# first turn the date columns
# into date types in R using
# the lubridate package
library(lubridate)
melted_cormat$x <- ymd(melted_cormat$x)
melted_cormat$y <- ymd(melted_cormat$y)
# then arrange the dataset by x and y dates
melted_cormat %>% arrange(x,y) -> melted_cormat Our final formatted matrix for plotting, called melted_cormat, looks like this (first 10 rows shown):
| x | y | count | meaningful_diff | |
|---|---|---|---|---|
| 1 | 2015-03-18 | 2015-03-18 | 0 | No Difference |
| 2 | 2015-03-18 | 2015-03-29 | 222 | No Difference |
| 3 | 2015-03-18 | 2015-05-12 | 737 | No Difference |
| 4 | 2015-03-18 | 2015-06-29 | 222 | No Difference |
| 5 | 2015-03-18 | 2015-07-19 | 515 | No Difference |
| 6 | 2015-03-18 | 2015-09-15 | 768 | No Difference |
| 7 | 2015-03-18 | 2015-09-22 | 884 | No Difference |
| 8 | 2015-03-18 | 2015-10-14 | 683 | No Difference |
| 9 | 2015-03-18 | 2015-10-18 | 837 | No Difference |
| 10 | 2015-03-18 | 2015-10-22 | 728 | No Difference |
The count variable here gives the number of times that the y date simulations were greater than the x date simulations. So the second row above indicates that the simulations on ‘2015-03-29’ were greater than those for ‘2015-03-18’ a total of 222 times.
We can make the heatmap with the following code:
# make the heatmap
ggplot(melted_cormat, aes(as.factor(x), as.factor(y),
fill = meaningful_diff)) +
# tile plot
geom_tile() +
# remove x and y axis titles
# rotate x axis dates 90 degrees
theme(axis.title.x=element_blank(),
axis.title.y=element_blank(),
axis.text.x = element_text(angle = 90,
vjust = .5)) +
# choose the colors for the plot
# and specify the legend title
scale_fill_manual(name = "Comparison",
values=c("#999999", "#800000")) +
# add a title to the plot
labs(title = "Pairwise Comparisons of Model-Estimated Step Counts at 6 PM") Which gives us the following plot:
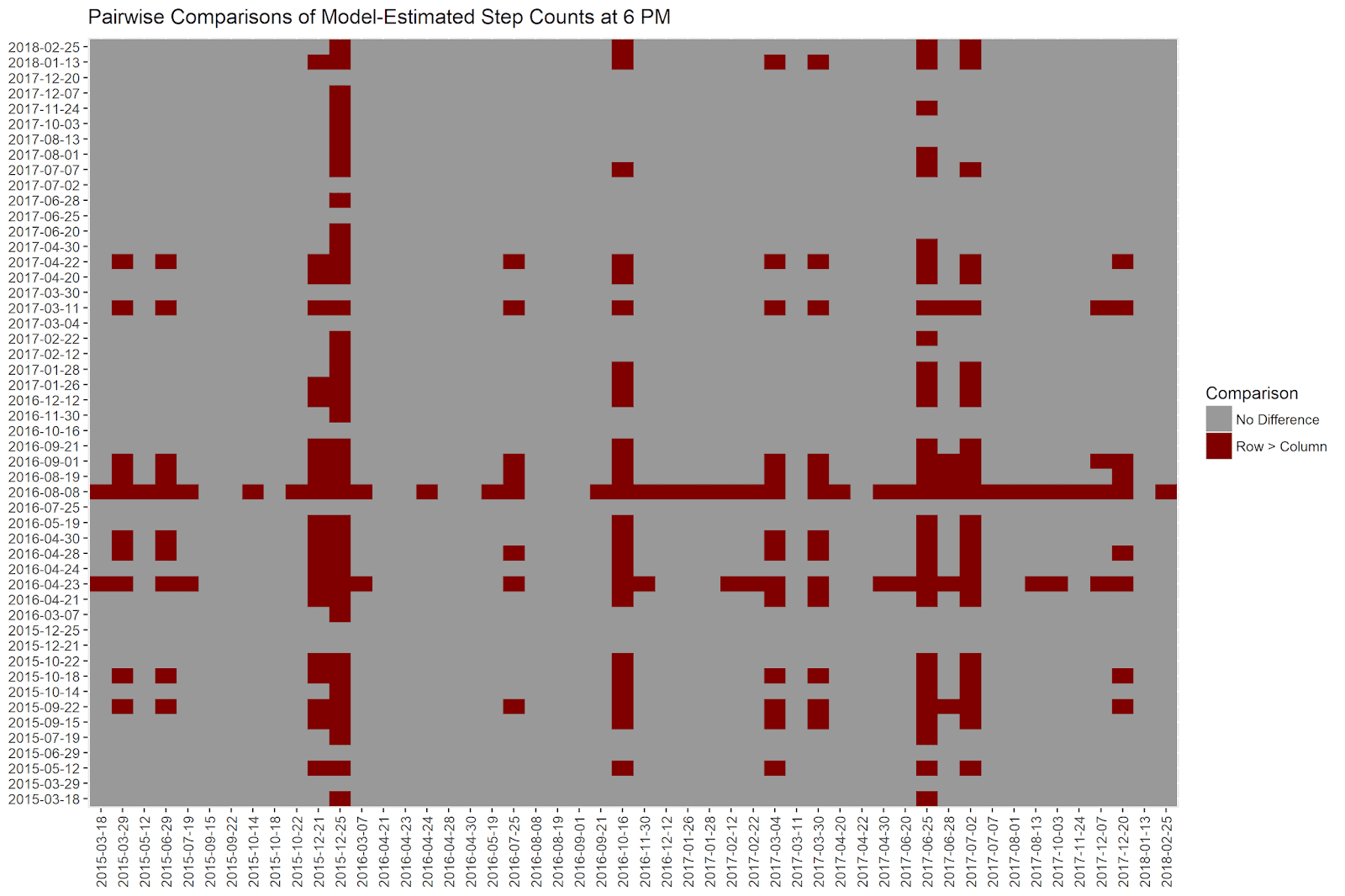
How can we read this plot? All of our 50 dates are displayed on the x and y axes. When the row step count is meaningfully larger than the column step count (according to our method, e.g. if more than 950 of the simulations for the row date are greater than those for the column date), the cell is colored in maroon. Otherwise, cells are colored in grey.
Rows that have a lot of maroon values are days that have higher step counts. To take a concrete example, the row representing ‘2016-08-08’ has many red values. If we look at the estimated step count for that day on the first graph above, we can see that it has the highest predicted step count in the sampled dates - over 25,000! It therefore makes sense that the estimated step count on this day is “significantly” larger than those from most of the other dates.
Columns that have many red values are days that have especially low step counts. For example, the column representing ‘2015-12-25’ is mostly red. As we saw above, this date has the lowest predicted step count in our sample - under 5,000! It is no wonder then that this date has a “significantly” lower step count than most of the other days.
How Bayesian Am I?
I’m a newbie to Bayesian thinking, but I get the sense that Bayesian statistics comes in many different flavors. The approach above strikes me as being a little bit, but not completely, Bayesian.
That’s So Bayesian
This approach is Bayesian in that it uses posterior distributions of daily step counts to make the comparisons between days. The approach recognizes that observed daily step counts are just observations from a larger posterior distribution of possible step counts, and we make explicit use of this posterior distribution in the data analysis. In contrast, frequentist methods basically only make use of point estimates of coefficients and the standard errors of those estimates to draw conclusions from data.
Not So Fast!
There are some things about this perspective that aren’t so Bayesian. First, we use flat priors in the model; a “more Bayesian” approach would assign prior distributions to the intercept and the coefficients, which would then be updated during the model computation. Second, we use the point estimates of our coefficients to compute the estimated daily step counts. A “more Bayesian” approach would recognize that the coefficient estimates also have posterior distributions, and would incorporate this uncertainty in the simulations of daily step counts.
What Do I Know?
This is my current best understanding of the differences in Bayesian and frequentist perspectives as they apply to what we’ve done here. I’m reading Statistical Rethinking (and watching the accompanying course videos) by Richard McElreath (love it), and these differences are what I’ve understood from the book and videos thus far.
The method used in this blog post is a nice compromise for someone comfortable with frequentist use of multi-level models (like myself). It requires a little bit more work than just interpreting standard multilevel model output, but it’s not a tremendous stretch. Indeed, the more “exotic” procedures (for a Bayesian newbie) like assigning priors for the model parameters are not necessary here.
Summary and Conclusion
In this post, we used multilevel modeling to construct pairwise comparisons of estimated step counts for 50 different days. We computed the multilevel model, extracted the coefficients and sigma hat value, and computed 1000 simulations from each day’s posterior distribution. We then conducted pairwise comparisons for each day’s simulations, concluding that a day’s step count was “significantly” larger if 950 of the 1000 simulations were greater than the comparison day. We used a heat map to visualize the pairwise comparisons; this heat map highlighted days with particularly high and low step counts as being “significantly” different from most of the other days. This allowed us to conduct these comparisons without the expensive adjustments that come with lowering our significance threshold for multiple comparisons!
[Edit - if you’re interested, you can find the data and code for this blog post at this link. Comments, suggestions, or improvements are very welcome!]
Coming Up Next
In the next post, we will move away from classical statistics and talk about machine learning techniques. Specifically, we will use a type of deep learning model to automatically generate band names.
Stay tuned!
-
Please correct me if I’m wrong! ↩